Congratulations to Fiona Whelan (MSc) for publishing her first manuscript “The Evolution of the class A scavenger receptors” in BMC Evolutionary Biology. The scavenger receptors are an evolutionarily ancient family of proteins required for host defence and homeostasis but teasing apart their function and even their structure has been challenging. The goal of this manuscript was to use evolution as a guide to discover how the class A scavenger receptor family was formed and to identify regions of conservation and hence probable functional importance for future study. Phagocytic receptors such as the class A scavenger receptors are integral members of the innate immune response, which is conserved in all classes of life and after reproduction and nutrient acquisition is probably the major most fundamental requirement for survival.
There are essentially only four basic mechanisms of the innate immune system – agglutination (e.g. lectins), lysis/neutralization (e.g complement, antimicrobial peptides), phagocytosis (e.g. scavenger receptors), and pro-inflammatory signalling (e.g. the toll like receptors). The fact that these processes are ancient and have been so strongly preserved is a testament to their importance. Of these, phagocytosis is likely the most ancient process and was probably adapted from its original purpose of nutrient ingestion . One might hypothesize that phagocytosis was truly the genesis of the immune system since our single celled ancestors had to distinguish between “self” and “non-self” in order to distinguish between food and their own daughter cells. From there phagocytosis became essential to fundamental processes such as embryonic development, pathogen recognition, and homeostatic clearance of senescent cells. Without phagocytosis, the transition to more complicated life forms could not have occurred.
Although there have been excellent evolutionary analyses of the lectins, toll like receptors and complement pathways, very little is known about the evolution of the phagocytic receptors. The class A scavenger receptors are an excellent example of these multifunctional receptors as they are involved in both host defence and homestasis. Since the phagocytic receptors in general and the scavenger receptors in particular are a diverse group of proteins,it has been challenging to understand how members within a group are related. Indeed, the first goal of this manuscript was to definitively demonstrate that the members of the class A scavenger receptors, which had been grouped together based on a ragtag combination of ligand binding and some degree of amino acid similarity, were actually a family at all. Since we were able to trace a probable path of gene duplication and consequent functionalization, we are confident that the 5 members (SRAI/II, MARCO, SCARA3/4/5) are actually related. Interestingly the class A scavenger receptors may have acquired their long stalk like form with a single scavenger receptor cysteine rich domain (SRCR) around the time of the evolution of fish since, although SRCR domain can be found in invertebrates and single celled organisms, we could not find anything that resembled a modern class A scavenger receptor in any genomes of evolutionarily more ancient organisms such as jellyfish, lampreys and insects.
Because elucidating the function of the specific domains of the scavenger receptors has been so challenging (even the function of the SRCR domain is unclear), ultimately we want to use evolution as a guide to which domains are functionally important (i.e. conserved). In this regard we found that there is a common conserved region in the collagenous domain, which in the type member SRAI, is believed to be the ligand binding domain. In addition conserved domains were identified in the cytoplasmic tail and the coiled-coiled domain. Future experiments will be performed to determine if these domains are necessary for structure, expression, cellular localization or phagocytic function.
The best part of our paper is that it is Open Access so you can enjoy reading it in provisional form today at http://www.biomedcentral.com/1471-2148/12/227/abstract. If you have questions, comments or thoughts, please feel free to contact Dr. Dawn Bowdish at bowdish@mcmaster.ca or on her Google+ account. Dr. Bowdish is always interested in talking with undergraduate or graduate students interested in pursuing studies that use bioinformatics to answer questions about the scavenger receptors, phagocytosis, and structure/function relationships.
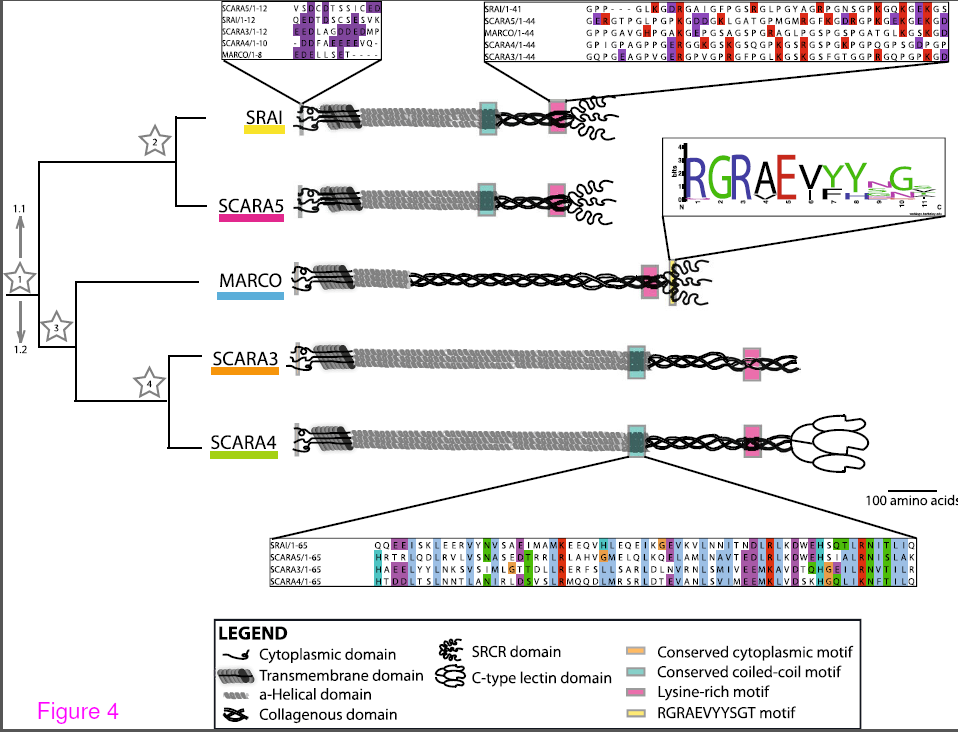
A summary of the common motifs in the class A scavenger receptor protein sequences. Conserved motifs present in the protein sequences of these receptors are indicated with coloured boxes at their approximate position within the protein with shout out boxes used to show the level of conservation across the aligned Homo sapiens sequences. From Whelan et al. BMC Evol Biol. BMC Evolutionary Biology 2012, 12:227