Bluesky Explainer thread by Dr. Kate Kennedy here:

Bluesky Explainer thread by Dr. Kate Kennedy here:

Link to publication here.
Link to Bluesky “Skeetorial” here and reproduced below without images:
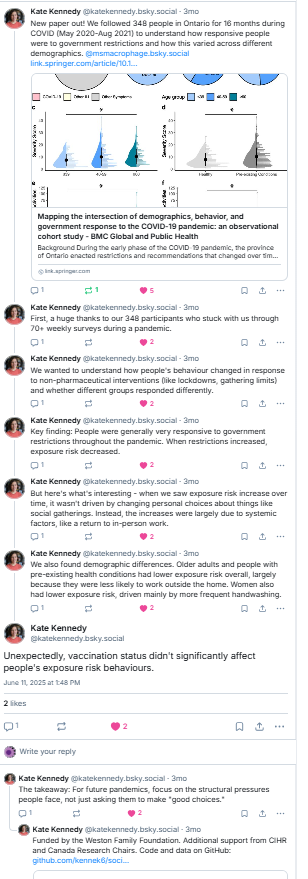
New paper! “Minimal Impact of Prior Common Cold Coronavirus Exposure on Immune Responses to Severe Acute Respiratory Syndrome Coronavirus 2 Vaccination or Infection Risk in Older Adults in Congregate Care”. For those of you who follow our #COVID work, read on for the story behind the story. 1/n
Remember reports like this one from the beginning of the pandemic? How could some older adults show such resilience to COVID compared to their peers? Some thought that they might have cross reactive immunity due to exposure to the related ‘seasonal’ or ‘common cold’ coronaviruses. 2/n
https://www.cbc.ca/news/canada/ottawa/102-year-old-woman-recovers-from-covid-19-1.5567189
After all, our @mcmasteru.bsky.social colleagues, Dr. Mark Loeb & team had shown years earlier that seasonal/common cold coronaviruses caused a lots of infections in long-term care and others had investigated whether these might protect kids from COVID…. 3/n
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0108481
….while others have shown that can be very deadly in residents of long-term care (reminding us that words matter and calling them ‘common colds’ minimizes risk – common viruses can still make people very sick, but that is rant for another day). So could pre-existing immunity be protective? 4/n
https://www.thelancet.com/journals/lanhl/article/PIIS2666-7568(23)00018-1/fulltext
Alternatively, maybe older adults got so sick because a life of exposure to these viruses ‘used up’ all the immune cells that could be used to respond to SARS-CoV-2 or COVID vaccines(i.e. ‘immune imprinting’, a phrase I came to hate along with ‘original antigenic sin’ as it was so misused) 5/n
To find out we tested whether antibody levels for the coronaviruses OC43, HL63, and 229E were higher/lower in people whose first COVID infection was an early Omicron variant and found they were not. Therefore it is unlikely these are either protective or problematic 6/n
What about pre-existing anti-coronavirus T cells? We looked at memory CD4+ and CD8+ T cells against the M & N proteins (indicates prev infections) and Spike (vaccine and infections). No evidence that these differed between those who did/did not get an infection (7/n)
What was a correlate of protection? Anti-RBD-IgG & neutralizing antibodies to the ancestral virus (which is all the residents would have been vaccinated to at that point). Unlike others we didn’t find that (serum) IgA was a correlate of protection. 8/n
Pre-existing immunity to common cold coronaviruses didn’t protect against SARS-CoV-2/COVID but might our vaccines and immunity alter immune responses to seasonal coronaviruses? Antibodies to other coronaviruses increased a bit (‘back-boosting’) after COVID infection or vaccination…. 9/n
But I doubt this will have much effect on the prevalence of other coronaviruses who follow a pretty consistent yearly/biennial or big wave/small wave pattern in the Northern hemisphere. We don’t know why but we do know that immunity doesn’t last long so a small boost from COVID infection/vaccination is not likely to make a difference 10/n
https://www.nature.com/articles/s41591-020-1083-1
Caveats: Only measured peoples first infections in the early Omicron era, only older adults living in LTC and retirement homes, vaccines would have been against the ancestral virus – things might be different in other populations/variants/vaccines. 11/n
Huge shout out to Braeden Cowborough for doing all those titres – that’s a lot of plates – and to Dr Jessica Breznik (former @miramcmaster.bsky.social currently @mcmasternexus.bsky.social PDF) for analytic skills. Thanks to the rest of the COVID-in-LTC team @mcmasteriidr.bsky.social
The Bowdish lab has received funding from the Canadian Institute for Health Research (CIHR) to study immune responses to SARS-CoV-2 infections and vaccinations in older adults including those living in long-term care. The applicant will use over 10,000 biobanked saliva, PBMCs, and serum from over 500 participants in addition to participant metadata and detailed infection records to;
The successful applicant will have significant leeway to develop an independent research project based on their interests and expertise. The applicant will work closely with an experienced technician and a graduate student, external collaborators, and will have access to biostatistics expertise. If interested, there will be opportunities to teach undergraduate courses.
The initial
contract will be for one year, with the possibility of extending for more
years, depending on productivity. The position is funded, but the applicant
will also apply for internal and external funding sources.
The Bowdish Lab is situated at Canada’s most research-intensive university, McMaster University in Hamilton, Ontario Canada. We are a diverse group of undergraduate students, graduate students, technicians and post-doctoral fellows committed to uncovering how the aging immune system changes and understanding why this alters immune responses to vaccination and respiratory infection. For more details on our lab’s philosophy see http://www.bowdish.ca/lab/lab-philosophy/ . We emphasize teamwork, career development, and leadership. Many of our former PDFs have gone on to independent faculty positions in Canada and abroad.
| Must have;Passion for discovery & evidence of leadership in the form of first-author publications in a relevant field.Team-player with history of collaboration and mentorshipFlow cytometry experienceDeep knowledge of immunity | Nice to have;Experience in human immunology/vaccinologyExperience in antibody quantitation and functional antibody assays (e.g., ADCC, ADP)Experience in intracellular cytokine staining or Experience in science communication with vulnerable populations. |
Please send Dr. Bowdish (bowdish@mcmaster.ca);
Click here to access: Monocyte-driven inflamm-aging reduces intestinal barrier function in females published in Immunity and Ageing, September 2024.
This publication by former PDF, Dr Candice Quin and team, discovers that inflammatory markers and gut permeability increase with age, but the leaky gut seems to be a female specific phenomena in both mice and humans.
Bluesky explainer thread below and here https://bsky.app/profile/msmacrophage.bsky.social/post/3l5ethyexgp2u
New publication alert! “Monocyte-driven inflamm-aging reduces
intestinal barrier function in females” by lead author Dr Candice Quin @uniofaberdeen.bsky.social. Read on for some surprising insights into sex differences in aging, the microbiome, inflammation, and the ‘leaky gut’ hypothesis….1/n
With age levels of inflammatory mediators (cytokines, CRP, & others) increase in the blood and tissues. This is often called ‘inflamm-aging’, and higher than age-average levels of these mediators are associated with chronic disease, frailty, and other age-associated ills. 2/n
Men & women age differently (‘men die quicker, women live sicker’)so it stands to reason they might ‘inflam-age’ differently, but very few studies on aging investigate sex differences in the aging trajectory. We looked at cellular & soluble inflammatory markers and saw sex differences! 3/n
We were surprised by the magnitude of sex differences but immunology is literally one of the worst disciplines when it comes to reporting by sex https://elifesciences.org/articles/70817
By why do we ‘inflamm-age’ in the first place? Might the mechanisms of inflamm-aging differ by sex 4/n
One of the major theories is that with age a dysbiotic microbiota causes the gut to become leaky (alternatively, the gut gets leaky with age and this alters the microbiota). Bacterial products leak out and cause inflammation. We’ve published in mice, others in other model organisms 5/n https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(17)30112-9?elsca1=etoc&elsca2=email&elsca3=1931-3128_20170412_21_4_&elsca4=Cell%20Press
Evidence for the ‘leaky gut’ hypothesis is weak in healthy humans (despite what the probiotic industry might say). Leakiness is generally found in people who have comorbidities, or are frail so we looked at healthy/non-frail male (41) females (54) btw 20-102 yrs and were surprised to find…6/n
…that women appeared to have leakier guts over the life course (as measured by the serum marker zonulin- imperfect), and only women had evidence of an age-associated increase in the bacterial product LPS in their serum. Could the leaky gut hypothesis be female specific? 7/n
Human studies are observational so we turned to mice to understand the chain of causality. Old female, but not male, mice did indeed have leakier guts and this seemed to be caused by TNF produced by the elevated number of circulating inflammatory monocytes. 8/n
Tl;dr The age-related leaky gut -> increased circulating bacterial products -> systemic inflammation hypothesis may only be true in females! (also – always investigate sex in your research) 9/n
Why? Well the female gut changes dramatically during pregnancy to increase nutrient absorption – perhaps that is why women seem to have more permeability over the life-course. We are doing some cool ongoing studies in vitro looking at male vs female monocytes ability to break the gut barrier. 10/n
Caveats: This was a non-frail, mostly white population – aging & immunology studies show tremendous differences by location so needs to be reproduced. Gut permeability measures were done by looking at serum markers – easy on the participant but indirect at best. 11/n
Thanks to the whole (unfortunately not on Bluesky team)! This was an incredibly fun and challenging project to work on but has cemented my commitment to considering sex in all the research we do. 👩🔬🧪🦠🚨 12/12
Download PDF here.
Link to journal here.
Abstract: Somatic mutations in the TET2 gene occur more frequently with age, imparting an intrinsic hematopoietic stem cells (HSCs) advantage and contributing to a phenomenon termed clonal hematopoiesis of indeterminate potential (CHIP). Individuals with TET2-mutant CHIP have a higher risk of developing myeloid neoplasms and other aging-related conditions. Despite its role in unhealthy aging, the extrinsic mechanisms driving TET2-mutant CHIP clonal expansion remain unclear. We previously showed an environment containing tumor necrosis factor (TNF) favors TET2-mutant HSC expansion in vitro. We therefore postulated that age-related increases in TNF also provide an advantage to HSCs with TET2 mutations in vivo. To test this hypothesis, we generated mixed bone marrow chimeric mice of old wild-type (WT) and TNF–/– genotypes reconstituted with WT CD45.1+ and Tet2–/– CD45.2+ HSCs. We show that age-associated increases in TNF dramatically increased the expansion
of Tet2–/– cells in old WT recipient mice, with strong skewing toward the myeloid lineage. This aberrant myelomonocytic advantage was mitigated in old TNF–/– recipient mice, suggesting that TNF signaling is essential for the expansion Tet2-mutant myeloid clones. Examination of human patients with rheumatoid arthritis with clonal hematopoiesis revealed that hematopoietic cells carrying certain mutations, including in TET2, may be sensitive to reduced TNF bioactivity following blockade with adalimumab. This suggests that targeting TNF may reduce the burden of some forms of CHIP. To our knowledge, this is the first evidence to demonstrate that TNF has a causal role in driving TET2-mutant CHIP in vivo. These findings highlight TNF as a candidate therapeutic target to control TET2-mutant CHIP.
Bluesky explainer thread here:
New Paper Alert: “Chronic TNF in the aging microenvironment exacerbates Tet2 lost-of-function myeloid expansion” published in Blood Advances #ImmunoSky #CHIP https://authors.elsevier.com/sd/article/S2473952924003860 (1/n)
Over time the progenitor cells (#stemcells) in our bone marrow acquire random mutations, with some genes being more likely to acquire these mutations than other. The Tet2 gene is one that acquires mutations. This is a problem because it regulates other genes through methylation (2/n)
Hematopoetic #stemcells with Tet2 mutations favour production of myeloid cells (#neutrophils #monocytes) over lymphoid cells (#Tcells #Bcells) and can outcompete stem cells that don’t have Tet2 mutations.(3/n)
If too many white blood cells originate from these mutants it is called Clonal Hematopoesis of Indeterminate Potential or #CHIP. CHIP is associated with an increase in mortality, and pneumonia (see prev paper), because immune cells with these mutations don’t work as well (4/n) https://shorturl.at/JNk6d
With age the proportion of Tet2 mutant cells increases and it is thought that the increased #inflammation that occurs with age contributes. Darwinian evolution happens in the bone marrow and stem cells with Tet2 mutations can outcompete (i.e. replicate more, make more white blood cells) than those without (5/n)
We investigated whether Tet2 mutant stem cells could outcompete others in the presence of the inflammatory cytokine TNF, which increases with age. We put a mix of normal and Tet2-/- stem cells into old mice that had lots of TNF and those with none (TNF KO). 6/n
Stem cells with Tet2 mutations could outcompete normal cells in old mice that had lots of TNF but not in TNF knockout mice. The aging inflammatory microenvironment contributes to CHIP! (7/n)
Cool finding- People with newly diagnosed #rheumatoidarthritis have lots of TNF and had low level of Tet2 and other CHIP mutants that went down as they started anti-TNF or anti-inflammatory therapy! Proof this happens in humans too (8/n)
Huge thank you to not on Bluesky Drs Michael Rauh (Queens), Candice Quin ( @uniofaberdeen.bsky.social ), Maggie Larche (UCalgary), Salman Basrai&Sagi Abelson @oicr.bsky.social and team. (9/9)
In this podcast first author Dr. Jessica A. Breznik of McMaster University, discusses the recently published manuscript titled “Diet-induced obesity alters intestinal monocyte-derived and tissue-resident macrophages and increases intestinal permeability in female mice independent of tumor necrosis factor.”
NEW & NOTEWORTHY We found that diet-induced obesity in female mice has tissue- and time-dependent effects on intestinal paracellular permeability as well as monocyte-derived and tissue-resident macrophage numbers, surface marker phenotype, and intracellular production of the cytokines IL-10 and TNF. These changes were not mediated by TNF.
Article Citation: Diet-induced obesity alters intestinal monocyte-derived and tissue-resident macrophages and increases intestinal permeability in female mice independent of tumor necrosis factorJessica A. Breznik, Jennifer Jury, Elena F. Verdú, Deborah M. Sloboda, and Dawn M. E. Bowdish
American Journal of Physiology-Gastrointestinal and Liver Physiology 2023 324:4, G305-G321
This CIHR funded PDF position will study how aging, and specifically age-associated inflammation alters myeloid cell development and macrophage function. This will include performing flow cytometry assays to quantitate development and maturation of myeloid cells in human blood and depending on the applicant’s interest and aptitude may include animal models. The successful applicant will be expected to develop a research project including all required experimental optimization, liaise with collaborators and research participants from multiple sights and write manuscripts to communicate research findings. The applicant must have a background in immunology and be a team player who is willing to mentor junior trainees and be an active participant in departmental seminars and events.
Essential skills
Flow cytometry
• This project requires significant skills in flow cytometry. Although most of the work will be done in human blood, expertise in mouse models would also be an asset. Please include details of your flow cytometry experience in your cover letter and be prepared to discuss the details of protocol development, trouble shooting and optimization if you are chosen for an interview.
Communication
• The applicant will be required to work closely with our research participants, including obtaining consent and filling out detailed health questionnaires. This will require the ability to describe the research in lay terms and to work with older adults who may have issues with hearing and site. The applicant will also be required to liaise with research co-ordinators from multiple sites to facilitate shipments and answer technical questions. Excellent English skills are essential.
• The applicant will be expected to present research findings to the lay public, research coordinators, nurses and PIs and will need experience speaking to broad audiences. The applicant will be responsible for publishing manuscripts. Please describe your oral and written communication skills as well as your publication history (published and in preparation) in your cover letter.
Must be willing to learn:
Phlebotomy
• This successful applicant must be willing to take a phlebotomy course and take blood from our research participants.
BSL2 level blood processing
• The applicant will be handling human blood, including blood that may be infected with viruses and consequently will need to be committed to following sterile and safe practices.
GLP procedures
• All our human immunology work is performed in a GLP compliant laboratory and the successful applicant must be willing to work with all the required GLP procedures.
Immunology assays
• The applicant will perform ELISAs including multiplex ELISAs (e.g. Luminex).
Statistics/R programming language/Data visualization
• Analysis of complex datasets (e.g. multilinear regression) and the R programming language is essential.
Additional skills which may be an asset:
Animal models (e.g. chimeric bone marrow transplants)
• There will be opportunities to test hypotheses and models using the Preclinical Studies in Aging Laboratory (PSAL: www.psal.ca), Canada’s only aging mouse colony. Specifically, there are opportunities to study how the aging microenvironment alters myeloid development by performing heterochronic bone marrow chimeras.
Immunosenescence/Senescence
• Experience in immunosenescence (mice and humans) would be a strong asset.
Microbiome analysis
• Opportunities exist to collaborate on projects on how the aging immune system alters the upper respiratory tract and gut microbiota. Experience with analysis of 16s rRNA sequencing, statistics and large dataset visualization would be an asset.
Please describe your experience with any of these techniques in your cover letter.
Why the Bowdish lab?
Our lab’s core values are Diversity, Ambition, Innovation and Collaboration. These core values dictate our approach to doing science. We support our trainees career development for careers both inside and outside of academia and this project will provide skills that will be broadly desirable no matter what the career trajectory. The Bowdish lab supports scientists with families and diverse backgrounds (see our lab’s diversity statement at http://www.bowdish.ca/lab/lab-philosophy).
If you are interested in applying contact Dr. Bowdish at bowdish@mcmaster.ca. Please include a coverletter detailing your research interests ambitions and relevant skills and a c.v. that includes your publications and references.
The lab that plays together stays (late nights scienc-ing) together, which is why the Bowdish lab had our annual retreat at Zacada circus school. Here we got some very sore muscles and discussed our successes and challenges of the past year and what our goals and ambitious are for the following year. Go Team!
The Bowdish lab is looking for new members to join our team! We currently have an opening for a post-doctoral fellow and a graduate student.
The PDF will project will involve investigating how the upper respiratory tract microbiome changes with age and declining immune function. Applicants must have a strong publication record in the field of immunology, microbiology, systems biology or molecular biology and applicants eligible for PDF funding from http://fhs.mcmaster.ca/mgdfa/ are particularly encouraged to apply (see link for eligibility). Experience in analysis of the microbiome or statistics of large/complex datasets are assets. Please provide a c.v. and a cover letter detailing your interest in the lab that includes contact details for references.
The graduate student position will be studying why aging macrophages are less able to kill bacteria. Applicants interested in beginning studies in January, May or Sept 2018 will be considered. Students aiming to pursue a PhD are preferred but exceptional MSc applicants will be considered. Previous research experience is strongly preferred. Candidates must have relevant courses in molecular/cellular biology, biochemistry, immunology or microbiology. Please include a transcript, a cover letter outlining your previous research experience and a list of references. Foreign students must have a scholarship to be considered.
Come join our strong team!